pycrtools.core.workspaces¶
| BoolArray | |
| BoolVec | c++ standard template library (STL) vector of type ‘bool’ |
| CRAverageSpectrumWorkSpace([parent, modulename]) | Workspace for mod::hCRAverageSpectrum. |
| CRFitBaselineWorkSpace([parent, modulename]) | Workspace for hCRFitBaseline. |
| CRMainWorkSpace([modulename]) | WorkSpace for global parameters. |
| CRQualityCheck(limits[, datafile, ...]) | Do a basic quality check of raw time series data, looking for rms, mean and spikes. |
| CRWorkSpace([parent, modulename]) | This class holds the arrays and vectors used by the various analysis tasks. |
| CRWorkSpace_default_doplot(self) | Make plots during processing to inspect data. |
| CRWorkSpace_default_t0(self) | The cpu starting time of the processingin seconds, used for benchmarking. |
| CRWorkSpace_default_verbose(self) | Print progress information during processing. |
| CRsetWorkSpace(ws, modulename, **keywords) | Sets the workspace in a function if not defined already and initialize parameters. |
| CheckParameterConformance(data, keys, limits) | Usage: |
| ComplexArray | |
| ComplexVec | c++ standard template library (STL) vector of type ‘complex’ |
| CoordinateTypes | |
| FFTWPlanManyDft | |
| FFTWPlanManyDftC2r | |
| FFTWPlanManyDftR2c | |
| FloatArray | |
| FloatVec | c++ standard template library (STL) vector of type ‘float’ |
| IntArray | |
| IntVec | c++ standard template library (STL) vector of type ‘int’ |
| StringArray | |
| StringVec | c++ standard template library (STL) vector of type ‘str’ |
| TBB2Data | |
| TBBData | |
| TYPE | |
| VecToString(self[, maxlen]) | see help of ‘hPrettyString’ |
| Vec_add(vec1, val) | Provides the + operator for adding two vectors or a vector and a scalar. |
| Vec_div(vec1, val) | Provides the / operator for dividing two vectors or a vector by a scalar. |
| Vec_iadd(vec1, vec2) | Provides the += operator for adding two vectors in place. |
| Vec_idiv(vec1, vec2) | Provides the /= operator for adding two vectors in place. |
| Vec_imul(vec1, vec2) | Provides the *= operator for addig two vectors in place. |
| Vec_isub(vec1, vec2) | Provides the -= operator for adding two vectors in place. |
| Vec_mul(vec1, val) | Provides the * operator for multiplying two vectors or a vector and a scalar. |
| Vec_neg(vec1) | Provides the - operator for a vector. |
| Vec_pos(vec1) | Provides the + operator for a vector (which is its identity: +vec = vec). |
| Vec_pow(vec1, val) | Provides the ** operator for raising a vector to a power. |
| Vec_rdiv(vec1, val) | Provides the / operator for dividing two vectors or a vector by a scalar. |
| Vec_rsub(vec1, val) | Provides the - operator for subtracting two vectors or a vector and a scalar. |
| Vec_sub(vec1, val) | Provides the - operator for subtracting two vectors or a vector and a scalar. |
| Vector([Type, size, fill, copy, properties]) | The basic Boost Python STL vector constructor takes no arguments and hence is a litte cumbersome to use. |
| asList(val) | Usage: |
| ashArray(val) | Usage: |
| asharray(self) | Return the argument as an hArray, if possible, otherwise as list. |
| asval(self) | Return the argument as a single value. |
| asvec(self) | Return the argument as a vector, if possible, otherwise as list. |
| atype | |
| basetype((IntVec) -> <type >) | basetype(FloatArray) -> <type ‘float’> |
| btype | str(object=’‘) -> string |
| commonpath(l1, l2[, common]) | |
| convert(fromvalue, totype) | Basis of a conversion routine, e.g. |
| extendflat(self, l) | Appending all elements in a list of lists to a one-dimensional vector with a flat data structure (just 1D). |
| fftw_flags | |
| fftw_sign | |
| get_filename(filename, ext) | Returns the folder name and its proper extention. |
| hArray([Type, dimensions, fill, name, copy, ...]) | Usage: |
| hArrayRead(filename[, block, restorevar, ...]) | Usage: |
| hArrayReadDictArray(dictionary, path[, ...]) | Recursively goes through a dict (of dicts) and replaces all placeholder (hFileContainer) with hArrays or Vectors read from disk. |
| hArrayWriteDictArray(dictionary, path, prefix) | Recursively goes through a dict (of dicts) and replaces all values which are hArray with a placeholder and writes the array to disk. |
| hArray_Find(self, operator[, threshold1, ...]) | Usage: |
| hArray_Select(self, *args, **kwargs) | Usage: |
| hArray_Set(self, value, *args, **kwargs) | Usage: |
| hArray_array(self) | array.array() -> hArray(array.vec,properties=array) |
| hArray_checksum(self) | array.checksum() -> Returns CRC32 checksum of a ‘list’ representation of the array |
| hArray_copy_resize(self, ary) | Retrieve the first element of the currently selected slice from the stored vector. |
| hArray_getHeader(self[, parameter_name]) | Usage: |
| hArray_getSlicedArray(self, indexlist) | self[n1,n2,n3]-> return Element with these indices |
| hArray_getinitargs(self) | Get arguments for hArray constructor. |
| hArray_getitem(self, indexlist[, asvec]) | ary[n1,n2,n3]-> return Element with these indices |
| hArray_getstate(self) | Get current state of hArray object for pickling. |
| hArray_hasHeader(self[, parameter_name]) | Usage: |
| hArray_list(self) | array.list() -> [x1,x2,x3, ...] |
| hArray_mprint(self) | ary.mprint() - > print the array in matrix style |
| hArray_new(self) | ary.new() -> new_array |
| hArray_newreference(self) | array.newreference() -> copy of array referencing the same vector |
| hArray_none(self) | array.none() -> None |
| hArray_par | Parameter attribute. |
| hArray_read(self, datafile, key[, block, ...]) | array.read(file,”Time”,block=-1) -> read key Data Array “Time” from file into array. |
| hArray_repr(self[, maxlen]) | |
| hArray_return_slice_end(val) | Reduces a slice to its end value |
| hArray_return_slice_start(val) | Reduces a slice to its start value |
| hArray_setHeader(self, **kwargs) | Usage: |
| hArray_setPar(self, key, value) | array.setPar(“keyword”,value) -> array.par.keyword=value |
| hArray_setUnit(self, *arg) | |
| hArray_setitem(self, dims, fill) | vec[n1,n2,..] = [0,1,2] -> set slice of array to input vector/value |
| hArray_setstate(self, state) | Restore state of hArray object for unpickling. |
| hArray_toNumpy(self) | Returns a copy of the array as a numpy.ndarray object with the correct dimensions. |
| hArray_toslice(self) | Usage: ary.toslice() -> slice(ary1,ary2,ary3) |
| hArray_transpose(self[, ary]) | Usage: |
| hArray_val(self) | ary.val() -> a : if length == 1 |
| hArray_vec(self) | array.vec() -> Vector([x1,x2,x3, ...]) |
| hArray_write(self, filename[, nblocks, ...]) | Usage: |
| hArray_writeheader(self, filename[, ...]) | Usage: |
| hCRAverageSpectrum(spectrum, datafile[, ws]) | Usage: |
| hCRCalcBaseline(baseline, frequency, ...[, ws]) | hCRCalcBaseline(baseline, coeffs, frequency,ws=None, **keywords): |
| hCRFitBaseline(coeffs, frequency, spectrum) | Function to fit a baseline using a polynomial function (fittype='POLY') or a basis spine fit to a spectrum while ignoring positive spikes in the fit (e.g., those coming from RFI = Radio Frequency Interference). |
| hFileContainer(path, name[, vector]) | Dummy class to hold a filename where an hArray is stored. |
| hNone2Value(none, defval) | Returns a default value if the the first input is the None object, otherwise return the value of the first argument. |
| hPlot_plot(self[, xvalues, xerr, yerr, ...]) | Method of arrays. |
| hSemiLogX(x, y, **args) | Total frustration avoid EDP64 crash on new Mac function |
| hSemiLogXY(x, y, **args) | Total frustration avoid EDP64 crash on new Mac function |
| hSemiLogY(x, y, **args) | Total frustration avoid EDP64 crash on new Mac function |
| hSliceListElementToNormalValuesEnd(s, dim) | |
| hSliceListElementToNormalValuesStart(s, dim) | |
| hSliceToNormalValues(s, dim) | Returns a slice object where none and negative numbers are replaced by the appropriate integers, given a dimension (length) dim of the full slice. |
| hVector_getinitargs(self) | Get arguments for hVector constructor. |
| hVector_getstate(self) | Get current state of hVector object for pickling. |
| hVector_list(self) | Retrieve the STL vector as a python list. |
| hVector_repr(self[, maxlen]) | Returns a human readable string representation of the vector. |
| hVector_setstate(self, state) | Restore state of hVector object for unpickling. |
| hVector_val(self) | Retrieve the contents of the vector as python values: either as a single value, if the vector just contains a single value, or otherwise return a python list. |
| hVector_vec(self) | Convenience method that allows one to treat hArrays and hVectors in the same way, i.e. |
| hWEIGHTS | |
| isVector(vec) | Returns true if the argument is one of the standard c++ vectors i.e. |
| ishArray((array) -> True or False) | Returns true if the argument is one of the hArray arrays, i.e. |
| listFiles(unix_style_filter) | Usage: |
| multiply_list(l) | Multiplies all elements of a list with each other and returns the result. |
| pathsplit(path) | This version, in contrast to the original version, permits trailing slashes in the pathname (in the event that it is a directory). |
| plot_draw_class(*args, **kwargs) | Just calls plt.draw - can be used in place of plotfinish in tasks to just plot and do nothing fancy |
| plotconst(xvalues, y) | Plot a constant line. |
| plotfinish([name, plotpause, doplot, ...]) | Usage: |
| readParfiles(parfile) | Open one or multipe parameter (i.e. |
| relpath(p1, p2) | |
| root_filename(filename[, extension]) | Will return a filename without the ending ”.pcr” |
| type2array((float) -> Vec(0)=[]) | Creates an array with elements of type ‘basetype’. |
| type2vector((float) -> Vec(0)=[]) | Creates a vector with elements of type ‘basetype’. |
| typename(btype) | basetype(float) -> “float” |
| v | |
| vtype | c++ standard template library (STL) vector of type ‘str’ |
| ws | Workspace for hCRFitBaseline. |
Provides a flexible data container class.
- class pycrtools.core.workspaces.CRAverageSpectrumWorkSpace(parent=None, modulename=None, **keywords)¶
Workspace for mod::hCRAverageSpectrum. See also CRMainWorkSpace and CRWorkSpace.
- default_blocks()¶
List of blocks to process.
- default_doplot()¶
Make plots during processing to inspect data.
- default_fft()¶
Array to hold the FFTed x-values (i.e. complex spectrum) of the raw time series data. (work vector)
- default_fx()¶
Array to hold the x-values of the raw time series data. (work vector)
- default_max_nblocks()¶
Absolute maximum number of blocks to average, irrespective of filesize.
- default_nblocks()¶
Number of blocks to average, take all blocks by default.
- default_t0()¶
The cpu starting time of the processingin seconds, used for benchmarking.
- default_verbose()¶
Print progress information during processing.
- class pycrtools.core.workspaces.CRFitBaselineWorkSpace(parent=None, modulename=None, **keywords)¶
Workspace for hCRFitBaseline. See also CRMainWorkSpace and CRWorkSpace.
- default_baseline_x()¶
Array holding the x-values and their powers for calculating the baseline fit.
- default_bwipointer()¶
Pointer to the internal BSpline workspace as integer. Don’t change!
- default_chisquare()¶
Returns the chisquare of the baseline fit. (output only)
- default_clean_bins_x()¶
Array holding the frequencies of the clean bins. (work vector)
- default_clean_bins_y()¶
Array holding the powers of the clean bins. (work vector)
- default_coeffs()¶
Polynomial coeffieients of the baseline fit. (output vector)
- default_covariance()¶
Array containign the covariance matrix of the fit. (outpur only)
- default_doplot()¶
Make plots during processing to inspect data.
- default_extendfit()¶
Extend the fit by this factor at both ends beyond numax and numin. The factor is relative to the unsued bandwidth.
- default_fftLength()¶
Length of unbinned spectrum.
- default_fittype()¶
Determine which type of fit to do: fittype=”POLY” - do a polynomial fit, else (“BSPLINE”) do a basis spline fit (default).
- default_freqs()¶
Array of frequency values of the downsampled spectrum. (work vector)
- default_height_ends()¶
The heights of the baseline at the left and right endpoints of the usable bandwidth where a hanning function is smoothly added.
- default_logfit()¶
Actually fit the polynomial to the log of the (downsampled) data. (Hence you need to .exp the baseline afterwards).
- default_meanrms()¶
Estimate the mean rms in the spectrum per antenna. (output vector)
- default_nbins()¶
The number of bins in the downsampled spectrum used to fit the baseline.
- default_ncoeffs()¶
Number of coefficients for the polynomial.
- default_nofAntennas()¶
Number of antennas held in memory.
- default_nselected_bins()¶
Number of clean bins after RFI removal. (output only)
- default_numax()¶
Maximum frequency of useable bandwidth. Negative if to be ignored.
- default_numax_i()¶
Channel number in spectrum of the maximum frequency of the useable bandwidth. Negative if to be ignored.
- default_numin()¶
Minimum frequency of useable bandwidth. Negative if to be ignored.
- default_numin_i()¶
Channel number in spectrum of the minimum frequency of the useable bandwidth. Negative if to be ignored.
- default_polyorder()¶
Order of the plyonomial to fit. (output only)
- default_powers()¶
Array of integers, containing the powers to fit in the polynomial. (work vector)
- default_ratio()¶
Array holding the ratio between RMS and power of the downsampled spectrum. (work vector)
- default_rms()¶
Array of RMS values of the downsampled spectrum. (work vector)
- default_rmsfactor()¶
Factor above and below the RMS in each bin at which a bin is no longer considered.
- default_selected_bins()¶
Array of indices pointing to clean bins, i.e. with low RFI. (work vector)
- default_spectrum()¶
Array of power values holding the downsampled spectrum. (work vector)
- default_t0()¶
The cpu starting time of the processingin seconds, used for benchmarking.
- default_verbose()¶
Print progress information during processing.
- default_weights()¶
Array of weight values for the fit. (work vector)
- default_xpowers()¶
Array holding the x-values and their powers for the fit. (work vector)
- class pycrtools.core.workspaces.CRMainWorkSpace(modulename=None, **keywords)¶
WorkSpace for global parameters.
Usage:
ws=CRMainWorkSpace()
This is a class to hold the variable, vectors, and arrays of all parameters used in the analysis.
You can access the parameters using ws["parametername"] and set them using ws["parametername"]=val.
Every known parameter has an associated function of the format .global_parameter to return a default value. The defaults will be set when calling the function ws.initParameters() or the first time when you access a parameter. A local copy will be made in an attribute of the class. So, you can access it also with:
ws.parametername
Workspaces can be stacked hierarchically in a tree structure, by providing a parent workspace as an argument during creation, e.g.:
child_ws=CRFitBaselineWorkSpace(ws)
Parameter can than be local or global. If a parameter is not yet set locally, it will be searched in the parent works space. If it is not found in either workspace, the default value will be calculated and assigned locally.
You can assign your own value with:
ws["parname"]=value
before initialization to avoid execution of the defaulting mechanism.
E.g. to set the blocksize to 1024, simply set ws["blocksize"]=1024 prior to calling initParameters.
Note that the local copy of a value is only made once initParameters is called or the parameter explicitly with ws["..."]. So, if a global value changes, the change will not be immediately reflected in the ws.parname value - hence use that with care.
To set all parameters (i.e. attributes) that do not exist yet, and assign a default value, use:
ws.initParameters()
A list of all parameters and their values are obtained with:
ws.help()
Available parameters:
Parameter Description nbins = 256 The number of bins in the downsampled spectrum used to fit the baseline. ncoeffs = 45 Number of coefficients for the polynomial. polyorder = 44 Order of the polynomial to fit. (output only) nofAntennas = 16 Number of antennas held in memory. freqs = hArray(float) Array of frequency values of the downsampled spectrum. (work vector) spectrum = hArray(float) Array of power values holding the downsampled spectrum. (work vector) rms = hArray(float) Array of RMS values of the downsampled spectrum. (work vector) rmsfactor = 2.0 Factor above and below the RMS in each bin at which a bin is no longer considered. verbose = True Print progress information during processing. selected_bins = hArray(int) Array of indices pointing to clean bins, i.e. with low RFI. (work vector) numax_i = 27459 Channel number in spectrum of the maximum frequency of the useable bandwidth. Negative if to be ignored. chisquare = Vec(int,16) Returns the 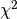 of the baseline fit. (output only),
e.g. [1,1,1,2,1,...]
of the baseline fit. (output only),
e.g. [1,1,1,2,1,...]doplot = False Make plots during processing to inspect data. numin = 12 Minimum frequency of useable bandwidth. Negative if to be ignored. ratio = hArray(float) Array holding the ratio between RMS and power of the downsampled spectrum. (work vector) xpowers = hArray(float) Array holding the 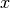 -values and their powers for the fit.
(work vector)
-values and their powers for the fit.
(work vector)bwipointer = 0 Pointer to the internal BSpline workspace as integer. Don’t change! nselected_bins = Vec(int,16) Number of clean bins after RFI removal. (output only), e.g. [236,236,239,239,238,...] clean_bins_y = hArray(float) Array holding the powers of the clean bins. (work vector) clean_bins_x = hArray(float) Array holding the frequencies of the clean bins. (work vector) baseline_x = hArray(float) Array holding the 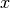 -values and their powers for calculating
the baseline fit.
-values and their powers for calculating
the baseline fit.numin_i = 3539 Channel number in spectrum of the minimum frequency of the useable bandwidth. Negative if to be ignored. covariance = hArray(float) Array containign the covariance matrix of the fit. (output only) numax = 82 Maximum frequency of useable bandwidth. Negative if to be ignored. logfit = True Actually fit the polynomial to the log of the (downsampled) data. (Hence you need to .exp the baseline afterwards). meanrms = Vec(float,16) Estimate the mean rms in the spectrum per antenna. (output vector) fftLength = 32769 Length of unbinned spectrum. height_ends = hArray(float) The heights of the baseline at theleft and right endpoints of the usable bandwidth where a hanning function is smoothly added. extendfit = 0.1 Extend the fit by this factor at both ends beyond  and
and  .
The factor is relative to the unsued bandwidth.
.
The factor is relative to the unsued bandwidth.t0 = 2.971779 The CPU starting time of the processingin seconds, used for benchmarking. weights = hArray(float) Array of weight values for the fit. (work vector) coeffs = hArray(float) Polynomial coeffieients of the baseline fit. (output vector) fittype = BSPLINE Determine which type of fit to do: fittype="POLY": do a polynomial fit. fittype="BSPLINE": do a basis spline fit (default). powers = hArray(int) Array of integers, containing the powers to fit in the polynomial. (work vector) - default_bad_channels()¶
Indexlist of bad channels containing RFI. (output only)
- default_baseline()¶
Array with a baseline fit to the spectrum.
- default_blocksize()¶
Size (number of values) of each block to be read in.
- default_cleanspec()¶
Copy of the spectrum with the gain curve and the spiky channels taken out.
- default_coeffs()¶
Polynomial coeffieients of the baseline fit. (output vector)
- default_datafile()¶
Datafile object. Will be created from the filename and set to the right blocksize, if it does not exist yet.
- default_doplot()¶
Make plots during processing to inspect data.
- default_fft()¶
FFT of the Raw time series antenna data.
- default_fftLength()¶
Size of the FFT or spectrum derived from the datareader object.
- default_filename()¶
Name of the data file to process.
- default_flaglist()¶
A list of bad antennas which failed the qualitycheck. (output only)
- default_frequency()¶
Frequency values (x-axis) corresponding to FFT and spectrum
- default_fx()¶
Raw time series antenna data.
- default_nbad_channels()¶
Number of bad channels (output only)
- default_ncoeffs()¶
Number of coefficients to describe the baseline.
- default_nofAntennas()¶
Number of antennas in the datafile (output only)
- default_numax()¶
Maximum frequency of useable bandwidth. Negative if to be ignored.
- default_numax_i()¶
Channel number in spectrum of the maximum frequency of the useable bandwidth. Negative if to be ignored.
- default_numin()¶
Minimum frequency of useable bandwidth. Negative if to be ignored.
- default_numin_i()¶
Channel number in spectrum of the minimum frequency of the useable bandwidth. (output only)
- default_qualitycriteria()¶
A Python dict with keywords of parameters and tuples with limits thereof (lower, upper). Keywords currently implemented are mean, rms, spikyness (i.e. spikyness).
Example:
qualitycriteria={"mean":(-15,15),"rms":(5,15),"spikyness":(-7,7)}
- default_rfi_nsigma()¶
Threshold for identifying a spike in Frequency as an RFI line to flag, in units of standard deviations of the noise.
- default_spectrum()¶
Power as a function of frequency.
- default_t0()¶
The cpu starting time of the processingin seconds, used for benchmarking.
- default_verbose()¶
Print progress information during processing.
- makeAverageSpectrum¶
alias of CRAverageSpectrumWorkSpace
- makeFitBaseline¶
alias of CRFitBaselineWorkSpace
- makeMain¶
alias of CRMainWorkSpace
- pycrtools.core.workspaces.CRQualityCheck(limits, datafile=None, blocklist=None, dataarray=None, nantennas=None, nbocks=None, maxblocksize=65536, blocksize=None, nsigma=5, verbose=True)¶
Do a basic quality check of raw time series data, looking for rms, mean and spikes.
Usage:
CRQualityCheck(limits,datafile=None,blocklist=None,dataarray=None,maxblocksize=65536,nsigma=5,verbose=True)
If a datafile is provided it will step through all (selected) antennas of a file, assess the data quality (checking first and last quarter of the file), and return a list of antennas which have failed the quality check and their statistical properties.
Instead of providing a datafile one can also provdide a data array, which will be processed in full.
Example:
>>> datafile=crfile(filename) >>> qualitycriteria={"mean":(-15,15),"rms":(5,15),"spikyness":(-7,7)} >>> flaglist=CRQualityCheck(qualitycriteria,datafile,dataarray=None,maxblocksize=65536,nsigma=5,verbose=True) # -> list of antennas failing the limits
Parameters:
Parameter Description qualitycriteria A Python dict with keywords of parameters and tuples with limits thereof (lower, upper). Keywords currently implemented are mean, rms, spikyness (i.e. spikyness). Example:
qualitycriteria={"mean":(-15,15),"rms":(5,15),"spikyness":(-7,7)}
datafile Data Reader file object, if None, use values in dataarray and don’t read data in again. datarray An optional data storage array to read in the data if no datafile is specified, this array should contain the data to inspect. In a pipeline where the function is called multiple times, it is recommended to always provide this array, since it saves one the creation and destruction of the array. blocksize The blocksize for reading in the data, will be determined automatically is not provided explicitly here. If None use information from datafile or use first dimension of dataarray. antennas How many antennas are in the dataarray or in the datafile. If None use information from datafile or use first dimension of dataarray. If equal to 0 then dataarray does not have that dimension. nblocks How many antennas are in the dataarray or in the datafile. If None use information from datafile or use first dimension of dataarray. maxblocksize If the blocksize is determined automatically, this is the maximum blocksize to use. blocklist The algorithms takes by default the first and last quarter of a file (and sets the blocksize accordingly). If you want to investigate all or other blocks, you need to provide the list explicitly here and also set the desired blocksize. nsigma Determines for the peak counting algorithm the threshold for peak detection in standard deviations. verbose Sets whether or not to print additional information.
- class pycrtools.core.workspaces.CRWorkSpace(parent=None, modulename=None, **keywords)¶
This class holds the arrays and vectors used by the various analysis tasks. Hence this is the basic workspace in the memory.
- setParameterDefault(par)¶
Assign a parameter its default value and make it local.
- setParameters(**keywords)¶
This method will set the parameters listed as arguments in the function call, i.e. modify the workspace attributes accordingly.
- pycrtools.core.workspaces.CRWorkSpace_default_doplot(self)¶
Make plots during processing to inspect data.
- pycrtools.core.workspaces.CRWorkSpace_default_t0(self)¶
The cpu starting time of the processingin seconds, used for benchmarking.
- pycrtools.core.workspaces.CRWorkSpace_default_verbose(self)¶
Print progress information during processing.
- pycrtools.core.workspaces.CRsetWorkSpace(ws, modulename, **keywords)¶
Sets the workspace in a function if not defined already and initialize parameters. One can provide a global workspace and the functions will pick the module corresponding to modulename
Parameters:
Parameter Description ws the workspace, if ws==None then create new one using function func modulename name of the (sub)module to use, will call the initialization function (naming convention: "CR"+modulename+"WorkSpace") if workspace does not exist. keywords local parameters to overwrite
- pycrtools.core.workspaces.CheckParameterConformance(data, keys, limits)¶
Usage:
qualitycriteria={“mean”:(-15,15),”rms”:(5,15),”spikyness”:(-3,3)}
CheckParameterConformance([Antenna,mean,rms,npeaks,spikyness],{“mean”:1,”rms”:2,”spikyness”:4},qualitycriteria) -> [“rms”,...]
Parameters:
data - is a list of quality values (i.e. numbers) to check
- keys - a dictionary of fieldnames to be checked and indices
- telling, where in data the field can be found
- limits - a dictionary of fieldnames and limits (lowerlimit,
- upperlimit)
Checks whether a list of numbers is within a range of limits. The limits are provided as a dictionary of fieldnames and tuples, of the form FIELDNAME:(LOWERLIMT,UPPERLIMIT). A list of fieldnames is returned where the data does not fall within the specified range.
- pycrtools.core.workspaces.hCRAverageSpectrum(spectrum, datafile, ws=None, **keywords)¶
Usage:
CRAverageSpectrum(spectrum,datafile,blocks=None,fx=None,fft=None)
Reads several blocks in a CR data file, does an FFT and then averages the powers to calculuate an average spectrum. The parameters are:
Parameters Description spectrum a float array of dimensions [nofAntennas,fftLength], containing the average spectrum. datafile a datareader object where the block size has been set appropriately. blocks a list of blocknumbers to read (e.g. range(number_of_blocks)). Default is to read all blocks. fx a work array of dimensions [datafile.nofAntennas,datafile.blocksize] which is used to read in the raw antenna data. Will be created if not provided. fft a work array of dimensions [datafile.nofAntennas,datafile.fftLength] which is used to calculate the FFT from the raw antenna data. Will be created if not provided. verbose Provide progress messages Available parameters in the Workspace (Examples):
DataReader object to read the data from:
datafile = crfile('/Users/falcke/LOFAR/usg/data/lofar/RS307C-readfullsecondtbb1.h5')
Absolute maximum number of blocks to average, irrespective of filesize:
max_nblocks = 3
Number of blocks to average, take all blocks by default:
nblocks = 3
List of blocks to process:
blocks = [0, 1, 2]
Print progress information during processing:
verbose = True
Array to hold the x-values of the raw time series data. (work vector):
fx = hArray(float)
Array to hold the FFTed x-values (i.e. complex spectrum) of the raw time series data. (work vector):
fft = hArray(complex)
The cpu starting time of the processing in seconds, used for benchmarking:
t0 = 2.971839
Make plots during processing to inspect data:
doplot = True
- pycrtools.core.workspaces.hCRCalcBaseline(baseline, frequency, numin_i, numax_i, coeffs, ws=None, **keywords)¶
hCRCalcBaseline(baseline, coeffs, frequency,ws=None, **keywords):
Calculate a smooth baseline from a set of coefficients that determine the baseline (e.g. as calculated by hCRFitBaseline) and an array of frequencies (which need not be equi-spaced).
- pycrtools.core.workspaces.hCRFitBaseline(coeffs, frequency, spectrum, ws=None, **keywords)¶
Function to fit a baseline using a polynomial function (fittype='POLY') or a basis spine fit to a spectrum while ignoring positive spikes in the fit (e.g., those coming from RFI = Radio Frequency Interference). The functions returns an array of coefficients of the polynomial or splines.
Use baseline.polynomial(frequency,coeffs,powers) to caluclate the baseline from the coefficients.
Parameters can be provided as additional keywords. e.g.:
baseline.polynomial(frequency,coeffs,powers, parameter1=value1, parameter2=value2)
or in a WorkSpace:
baseline.polynomial(frequency,coeffs,powers, ws=WorkSpace),
or in a mix of the two. Otherwise default values are used.
A full list of parameters can be obtained with with:
CRFitBaselineWorkSpace().help()
- pycrtools.core.workspaces.ws¶
alias of CRFitBaselineWorkSpace