Running the cosmic rays pipeline¶
This recipe explains briefly the LOFAR cosmic rays analysis pipeline from a practical users perspective
Introduction¶
This recipe assumes that you have a working installation of the LOFAR user software in the directory pointed to by the bash environment variable $LOFARSOFT. The recipe also assumes that you have an svn account for the LOFAR respository.
On the coma cluster the software is located in /vol/optcoma/lofarsoft. This directory contains a checkout from the subversion repository http://usg.lofar.org/svn/code/trunk and was build by a make pycrtools in the build subdirectory. To perform an update of the software simply run svn up in this directory and repeat the make.
The PyCRTools source code is located in the $LOFARSOFT/src/PyCRTools subdirectory and this recipe assumes that the $PYCRTOOLS environment variable points to this directory.
Important files and directories¶
The $PYCRTOOLS directory contains several files and directories that are important for the cosmic rays pipeline. They require a certain structure in your home folder, please check and change accordingly if needed.
- pipelines directory containing all pipeline scripts.
- pipelines/cr_physics.py main analysis pipeline.
- pipelines/cr_simulation.py simulation pipeline.
- scripts/run_crdb_populate.py crawls hdf5 files and adds new events to database.
- jobs directory containing job scripts for cronjob starting of pipelines.
- jobs/cr_pipeline.sh calls the following scripts:
- jobs/cr_physics.sh script to start analysis pipeline,
- jobs/cr_populate_database.sh script to populate database with new events.
- cr_simulations.sh script to start the simulation pipeline,
- extras/crserver directory containing the webserver scripts and layout files.
- extras/crserver/crserver.py the webserver itself.
Adding new events to the database¶
The raw LOFAR data is stored in .h5 files on the coma cluster in /vol/astro3/lofar/vhecr/lora_triggered/data. When new events are added to this directory, or when existing events are updated with data from stations that were previously unavailable, they are added to the database by a cronjob executing cr_populate_database.sh from the $PYCRTOOLS/jobs directory. This script in turn calls run_crdb_populate.py and add_beam_direction.py to crawl over all files and add changes to the database. The crontab entry (edited with crontab -e) looks like:
0 0 * * * sbatch -p short $PYCRTOOLS/jobs/cr_populate_database.sh &> /dev/null
Running the LORA analysis¶
The software that is used to reconstruct the LORA data, to such a format that it can be used by the pipeline, is located in the subversion repository hosted at https://svn.science.ru.nl/repos/lora_software. The actual analysis software is based on C++ and ROOT (CERN software) and was written by Satyendra Thoudam. In order to be run automatically by a cron job, the script runninglora_coma.py is used (also in the repository). The file lora.job (also in the repository) is a suggestion of what to run as a daily cronjob. The script will, when run, collect all relevant data from the coma cluster, generate input files, execute the analysis and copy the final data product to where the cosmic ray pipeline finds it.
Required are (besides a working installation of ROOT and PyCRtools as they are on coma):
- the raw LORA data, currently stored in /vol/astro3/lofar/vhecr/lora_triggered/LORAraw These data are copied by a cron job from the LORA master computer in the LOFAR field.
- In case corrupted filed are in this data-set, they need to be excluded in the first couple of lines in runninglora_coma.py in order to avoid crashes, when they are rsynced again.
- the trigger log called LORAtime4 stored in /vol/astro3/lofar/vhecr/lora_triggered/LORA. This file is copied by a cron job from the LOFAR cluster and is updated every night. (ask Sander ter Veen for details)
The first analysis step will generate calibrated data files per month, such as output_201501.root (to be found in the relative directory analysis/LORAOutput/).
The second parameterization step generates data files, such as LORAdata-20150114T205644.dat, LORAdata-20150114T205644.png, LORAdata-20150114T205644.eps. The .dat file is needed to fill the pipeline. The .png file will be displayed on the webserver. The .eps file is for publication purposes, if needed.
Updating and compiling¶
In order to run any scripts, “use offline” is required. This sets the ROOT paths to the ROOT version used in offline (software Pierre Auger Collaboration, in case of problems please coordinate with whoever is currently in charge). In case this needs to be updated the software needs to be recompiled with make main_analysis and make main_parameterization in the analysis folder. Note: make clean sometimes does not remove all files correctly so make sure to check.
Future needs¶
The software will need to be updated once:
- LORA is being expanded and gets more/different data
- the trigger file LORAtime4 changes format or does no longer exist
- the requirements of the LOFAR analysis pipeline change
How the pipeline deals with this data¶
The data files LORAdata- and LORAtime4 are read using the lora.py module in pycrtools. This also contains all the magic of correcting timeoffsets etc. The script “update_loradata.py” in the $PYCRTOOLS/scripts folder can be used to update LORA data, if the automated running has been cancelled for a while or if due to any reason the events are not filled with LORA data and are skipped by the pipeline.
Suggestions for rerunning¶
If a bug is found, all data needs to be regenerated. After that the database needs to be updated with the new parameters. Please use the options of runninglora_coma.py (LORA svn repository) to rerun the LORA data. It is wise to first regenerate all analysis files and then run the parameterization once over all data. Only after this, the database should be updated.
Running the analysis pipeline¶
The cosmic rays analysis pipeline consists of one top level Python script cr_physics.py and associated modules and tasks that are called from it. All results are written to the directory /vol/astro3/lofar/vhecr/lora_triggered/results and the database.
The pipeline is started on the cluster by having a cronjob execute the script cr_pipeline.sh. The crontab entry (edited with crontab -e) looks like:
0 3 * * * $PYCRTOOLS/jobs/cr_pipeline.sh > $HOME/cr_pipeline.log
This script determines how many events are marked with status NEW in the database and submits a single array job to the SLURM scheduler for which each task executes cr_physics.sh (which simply calls cr_physics.py with the correct parameters and environment). Thus, the easiest way to (re)process some or all events is to mark them as NEW in the database and run cr_pipeline.sh on coma.science.ru.nl.
Running the simulation pipeline¶
The simulation pipeline consists of one file cr_simulation.py. Simulations are first written locally into the /scratch/cr_sim_pipeline directory of the compute node where the script was executed. Then they are copied to /vol/astro3/lofar/sim/pipeline/events.
This pipeline is started on the cluster by having a cronjob execute the script cr_simulations.sh. The crontab entry (edited with crontab -e) looks like:
0 8 * * * $PYCRTOOLS/jobs/cr_simulations.sh > $HOME/cr_simulations.log
About the webserver¶
The webserver consists of a single Python script crserver.py should be running on the same server that contains the PostgreSQL database. It can be accessed by going to port $8000/events$ on that machine, in our case mainly http://crdb.astro.ru.nl:8000/events. (All events can be found at http://crdb.astro.ru.nl:8000/events ). General statistics (generated on the fly from the database) are available at http://crdb.astro.ru.nl:8000/statistics.
The webserver looks for figures in the /vol/astro3/lofar/vhecr/lora_triggered/results directory on the local machine and hence needs read access. It also expects write access to /vol/astro3/lofar/vhecr/lora_triggered/statistics to write the statistics plots.
When the webserver is not working or displaying things incorrectly, it probably needs to be restarted. This can only be done by C&CZ, so send a mail to the postmaster.
Adding or removing database parameters¶
This should never be needed for stable operation, but might be desired for new analysis. It is a simple procedure.
- First add the parameter by editing $PYCRTOOLS/modules/crdatabase.py and adding a new version to the end of the __updateDatabase function. Don’t forget to give it a higher version number and increasing the value of self.db_required_version, further up in the file, to match.
- Commit your changes to the lofar.usg repository.
- Then update the code on coma using the instructions from the introduction of this document.
- The webserver generates XML files from database queries, transforms them to HTML through XSL files and formats them using the CSS files in $PYCRTOOLS/crserver/layout. Mostly no changes are needed. Only when changing some of the top level parameters (e.g. the ones not in the *_parameters tables) is editing to both the queries and the XSL files needed. If changes have been made to the queries in crserver.py the webserver needs to be restarted.
That should be it! The database will be updated automatically when it is opened by the pipeline the next time.
What can go wrong¶
The http://crdb.astro.ru.nl:8000/statistics page is a good starting point to check how the events and simulations are doing. Here is a typical list of failures that might occur.
- An event will be in the state ERROR: The pipeline is designed as such that all crashes in running of cr_physics.py should be caught and are used to change the status of the event in the database to ERROR. The log-file of this event will give you an indication what has gone wrong. The plots will still be old, if the pipeline has run before, so they cannot be used to check. The rest is debugging the analysis pipeline.
- The LORA data is not present for events and they are SKIPPED. Reasons can be an corrupt datafile, which then needs to be excluded (see above) or an interruption of the data-transfer. In this case, rerun the LORA reconstruction and use the procedure as shown above to update the events. Note: Single events with skipped data occur from time to time. You should only be worried about several events with missing data in a row.
- Some one found a bug in the analysis. This probably means you will need to update PyCRTools and rerun the whole pipeline. This is easiest done by setting the status of all events to NEW as shown above.
- Simulation status is HOLD. This requires someone to inspect the data, as this is triggered by an unusual
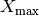 value or energy. The script ($PYCRTOOLS/scripts) simulation_holds.py can be used release the simulations with different reasons. This requires physics input and is a designed break point for the pipeline. If you don’t know what to do simply let the events stay on HOLD.
value or energy. The script ($PYCRTOOLS/scripts) simulation_holds.py can be used release the simulations with different reasons. This requires physics input and is a designed break point for the pipeline. If you don’t know what to do simply let the events stay on HOLD. - The database server died or someone managed to delete parts of the database. Please try not to do this! However, if this happens it does not mean that everything is lost. Delete all (if any) remaining tables from the database and start by adding the events again. The pipeline will need to be rerun and the simulation pipeline will hopefully figure out what it has and where to continue. There are, however, some manual entries, like flagged events and simulations that have been released, which can not be recovered if the database is lost.